
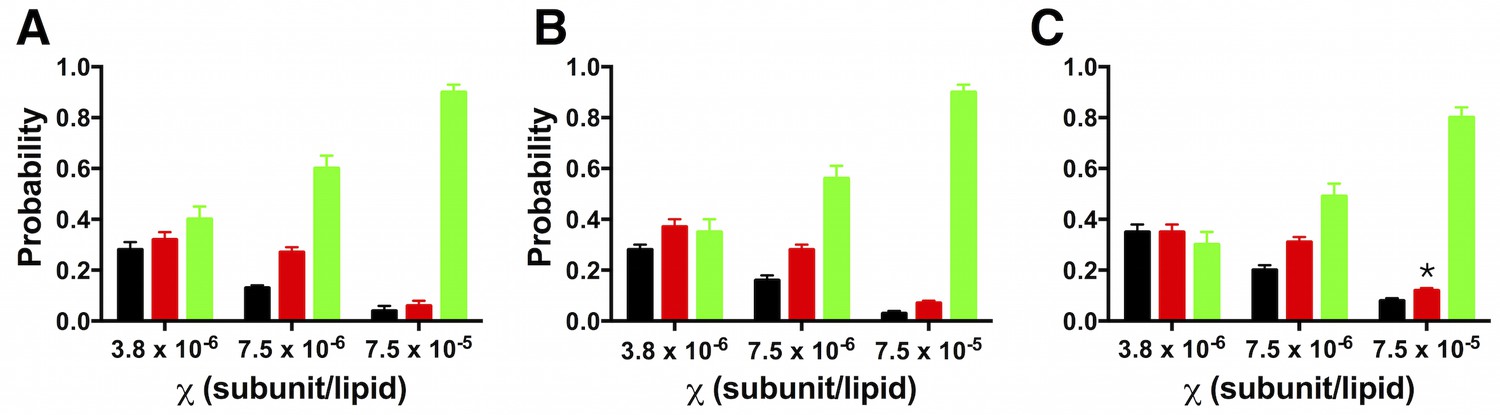
Furthermore, many genes involved in the flavonoid and phenylpropanoid pathways were highly expressed in Viroflay compared to Solomon at 168 hpi. The types of genes significantly up-regulated in Solomon in response to the pathogen suggests that salicylic acid and ethylene signaling pathways mediate resistance. In Solomon, the hypersensitive inducible genes such as pathogenesis-related gene PR-1, glutathione-S-transferase, phospholipid hydroperoxide glutathione peroxidase and peroxidase were significantly up-regulated uniquely at 48 hpi and genes involved in zinc finger CCCH protein, glycosyltransferase, 1-aminocyclopropane-1-carboxylate oxidase homologs, receptor-like protein kinases were expressed at 48 hpi through 168 hpi. Fold change differences in gene expression were recorded between the two cultivars to identify candidate genes for resistance. In this study, we took advantage of new spinach genome resources to conduct RNA-seq analyses of transcriptomic changes in leaf tissue of resistant and susceptible spinach cultivars Solomon and Viroflay, respectively, at an early stage of pathogen establishment (48 hours post inoculation, hpi) to a late stage of symptom expression and pathogen sporulation (168 hpi).
New pathotypes of this pathogen are increasingly reported which are capable of breaking resistance. The disease causes significant economic losses, especially in the organic sector of the industry where the use of synthetic fungicides is not permitted for disease control. Downy mildew of spinach is caused by the obligate oomycete pathogen, Peronospora effusa.


 0 kommentar(er)
0 kommentar(er)
